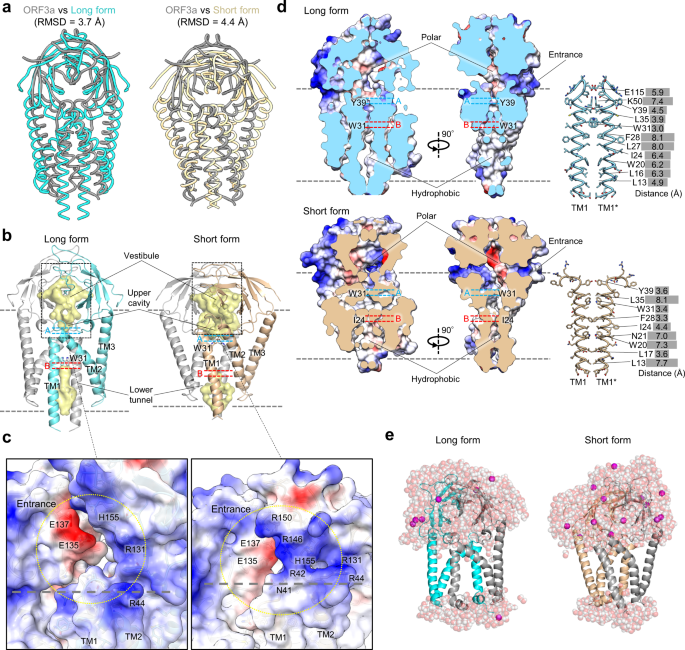
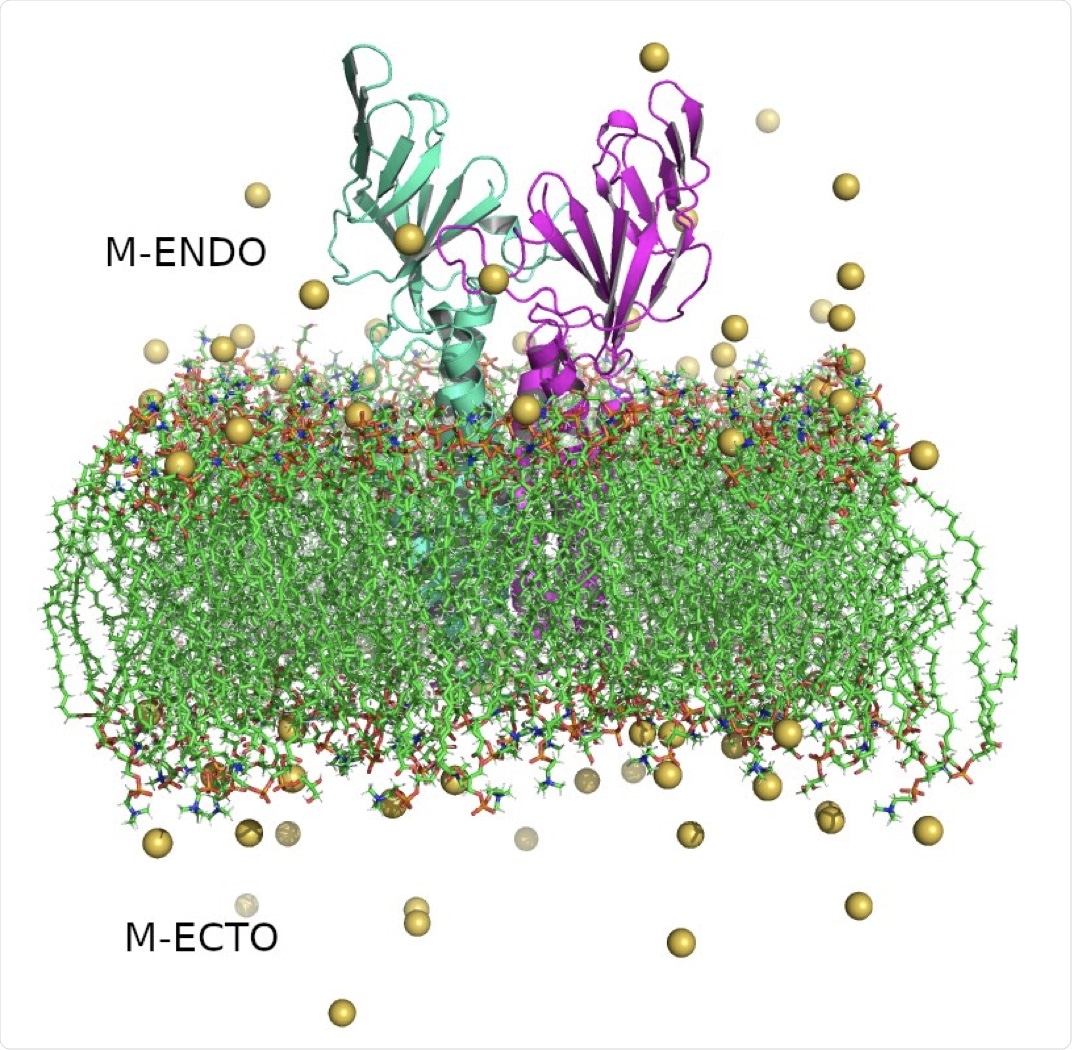
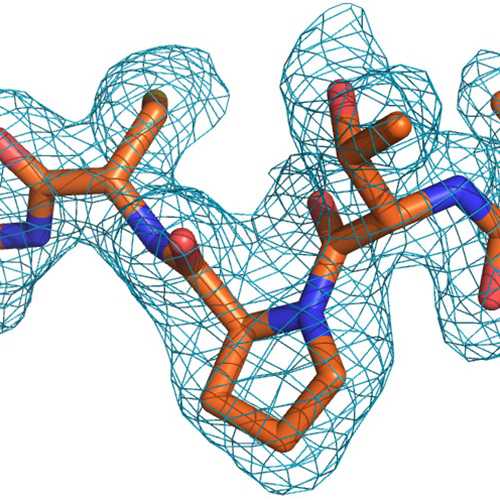
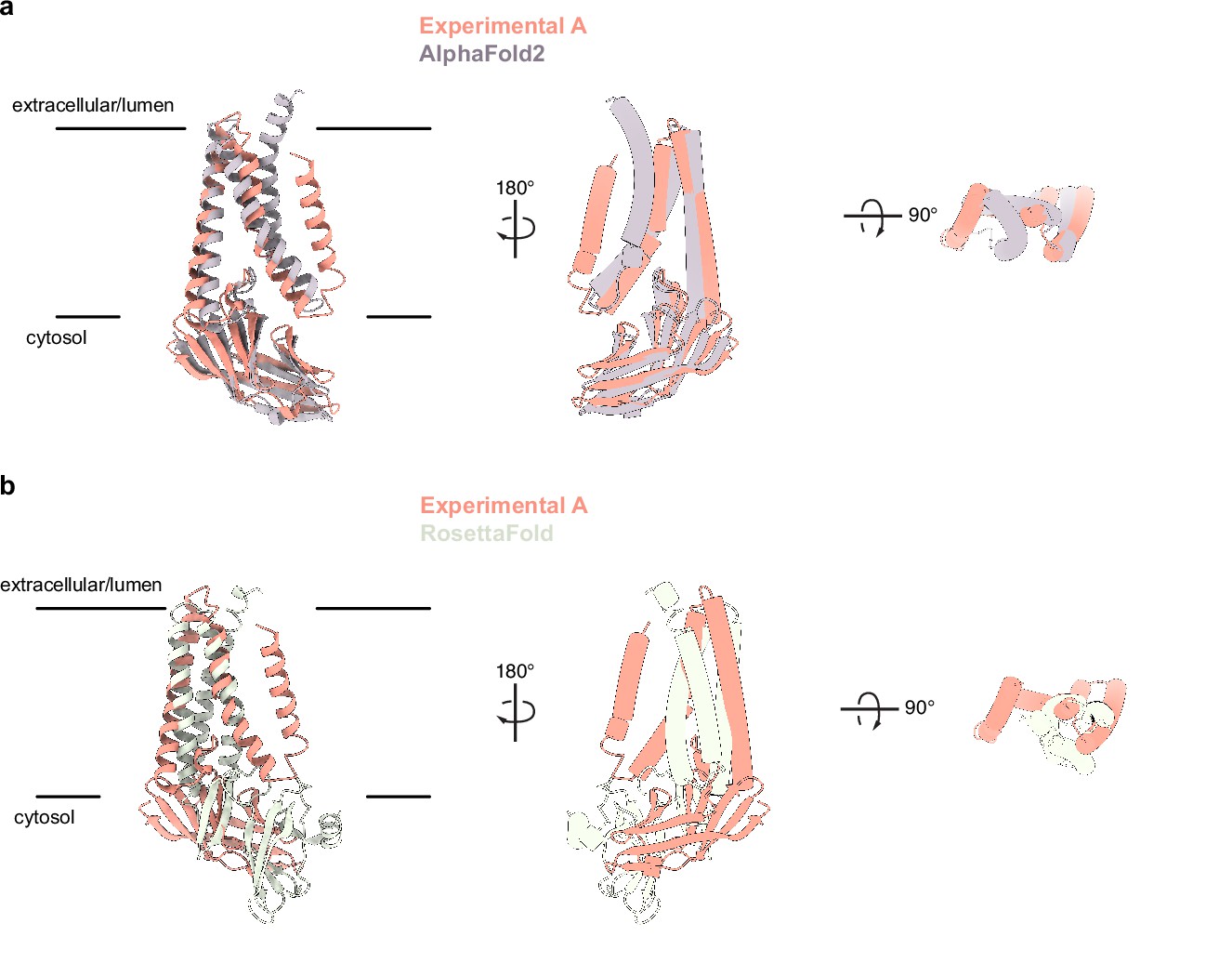
Probing the formation, structure and free energy relationships of M protein dimers of SARS-CoV-2 - ScienceDirect

Structural determination of Streptococcus pyogenes M1 protein interactions with human immunoglobulin G using integrative structural biology | bioRxiv

Deep Dive into Machine Learning Models for Protein Engineering | Journal of Chemical Information and Modeling

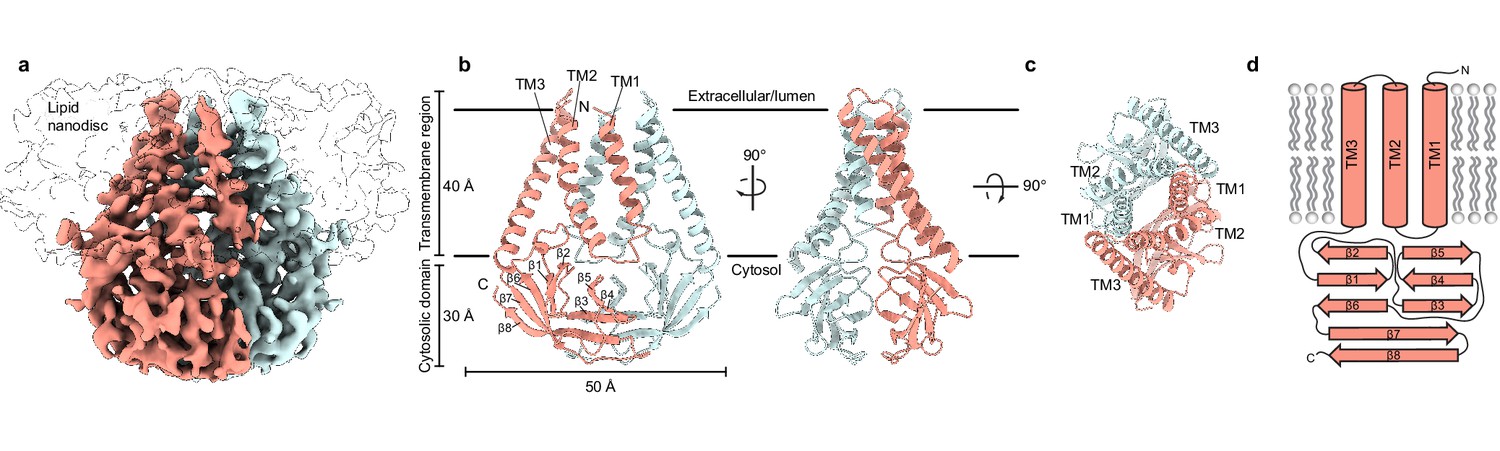
Structure and dynamics of the SARS‐CoV‐2 envelope protein monomer - Kuzmin - 2022 - Proteins: Structure, Function, and Bioinformatics - Wiley Online Library
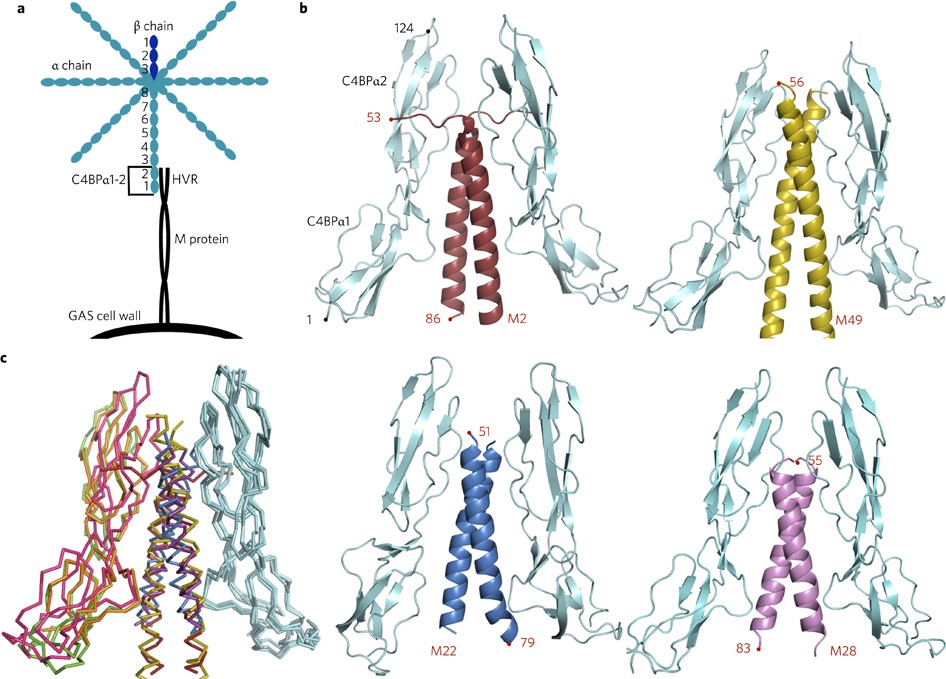
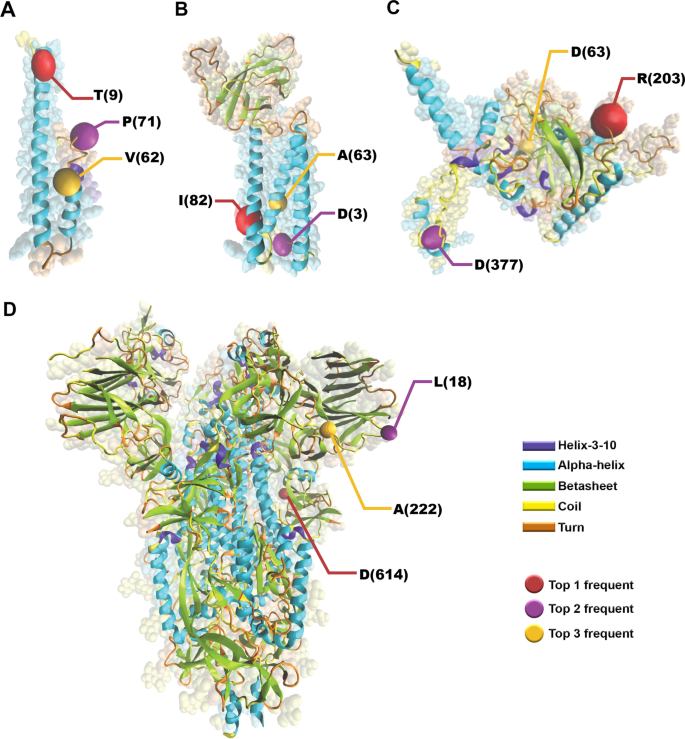
Conserved patterns hidden within group A Streptococcus M protein hypervariability recognize human C4b-binding protein | Nature Microbiology

3: Structure of the S. pyogenes M protein. (A) The primary structure of... | Download Scientific Diagram

Streptococcus pyogenes Forms Serotype- and Local Environment-Dependent Interspecies Protein Complexes | mSystems

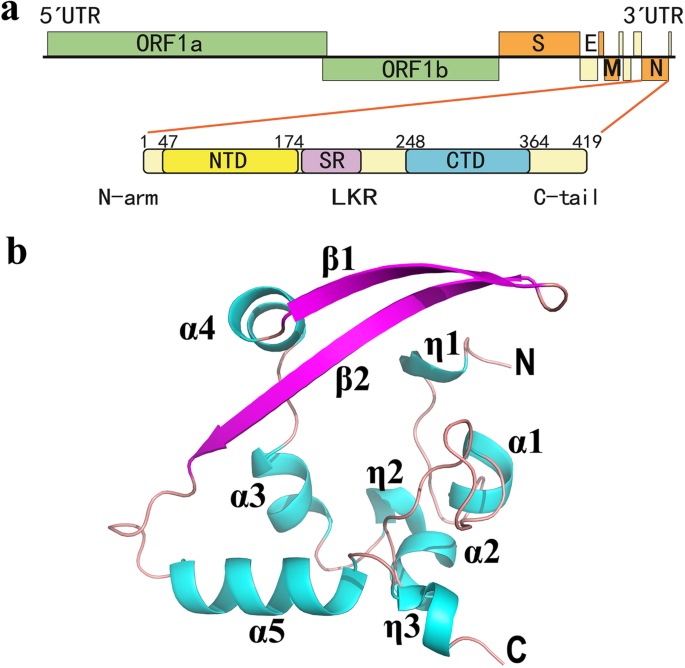
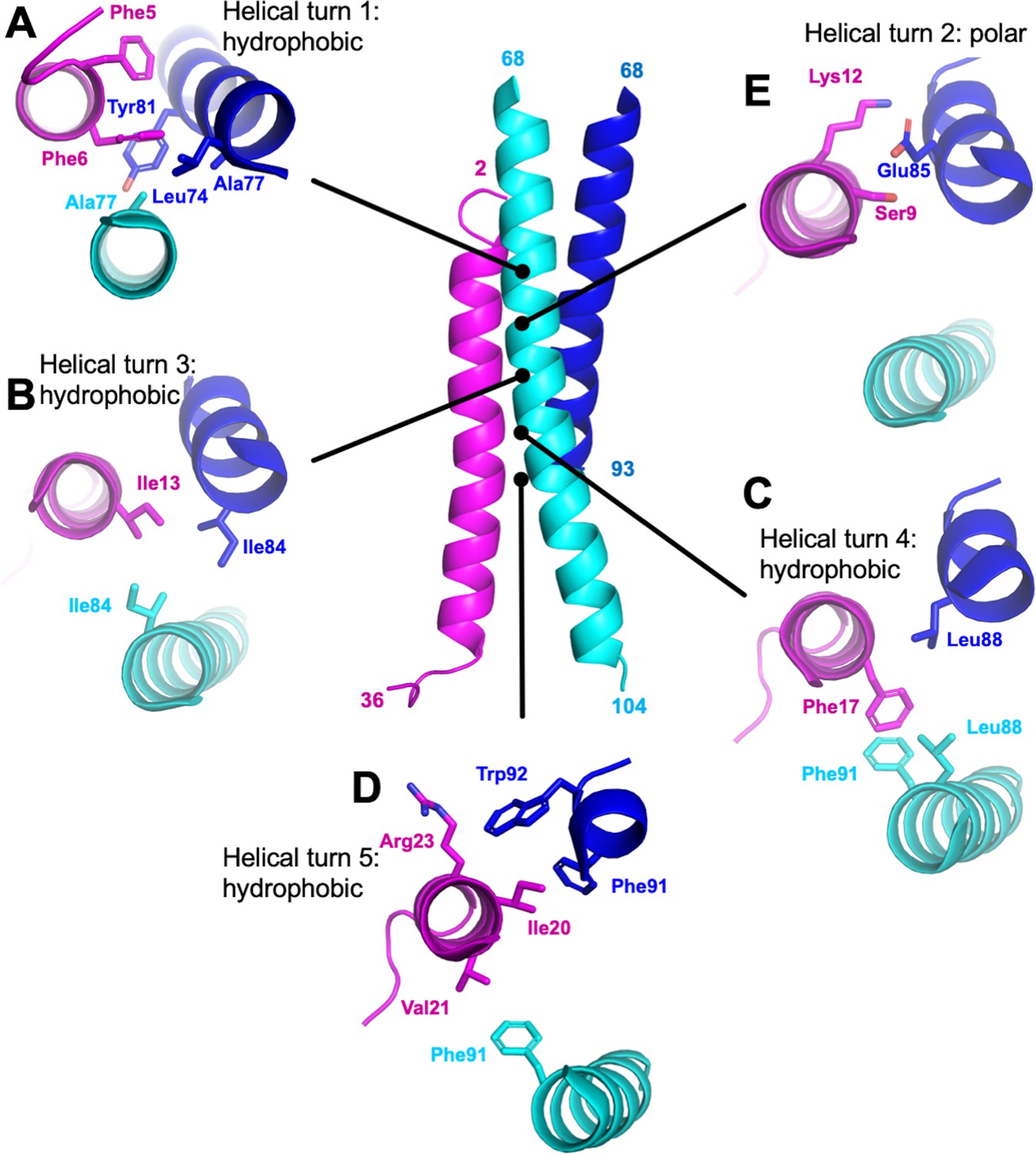
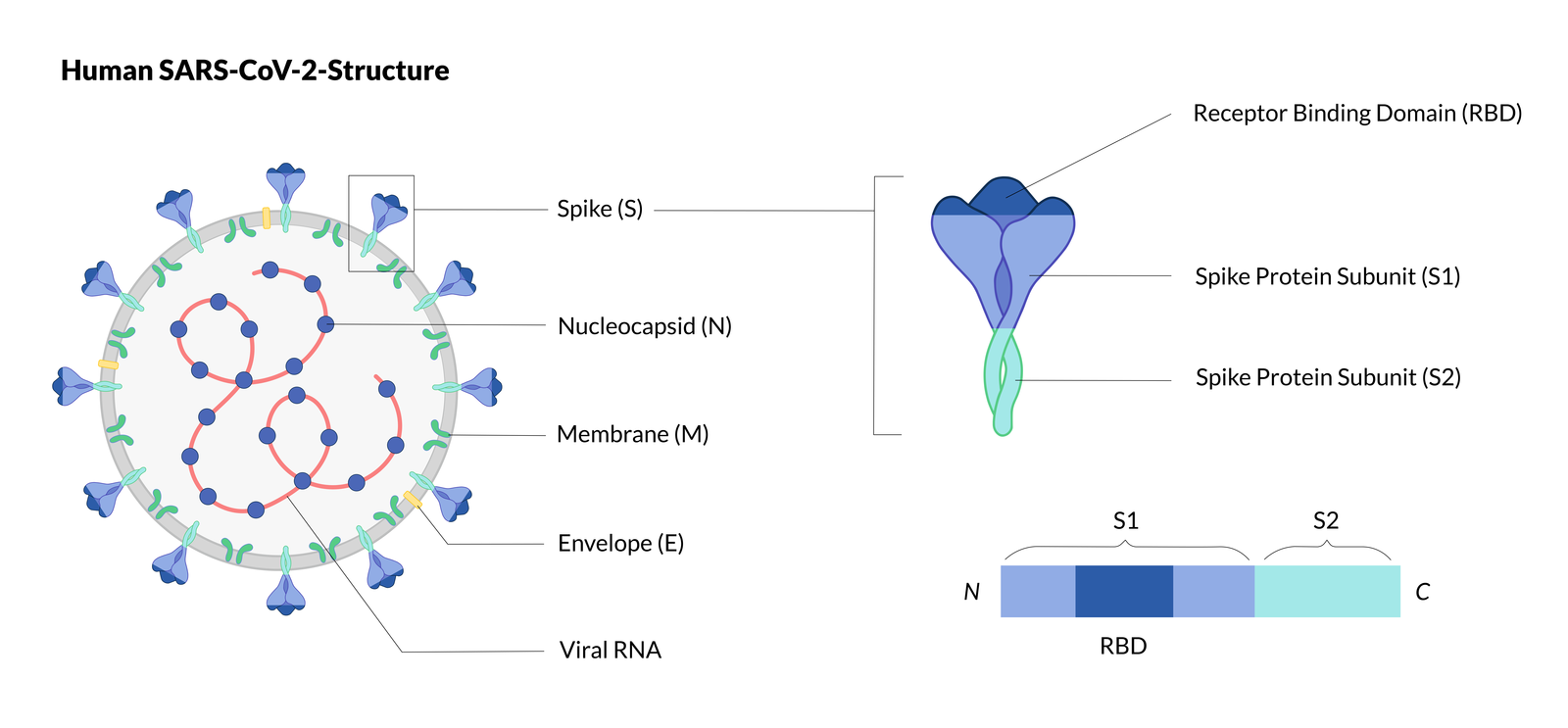
Architecture and self‐assembly of the SARS‐CoV‐2 nucleocapsid protein - Ye - 2020 - Protein Science - Wiley Online Library
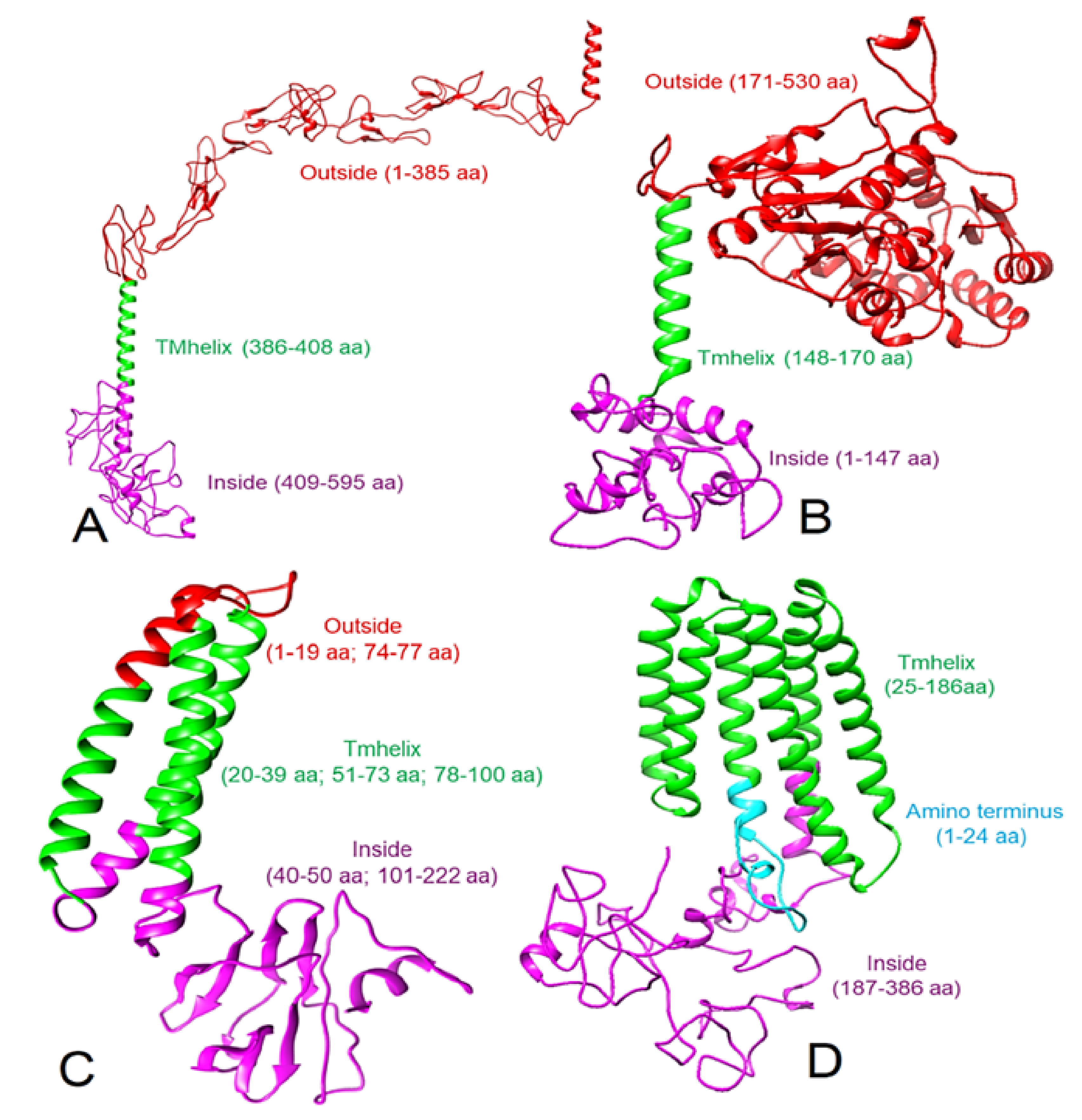
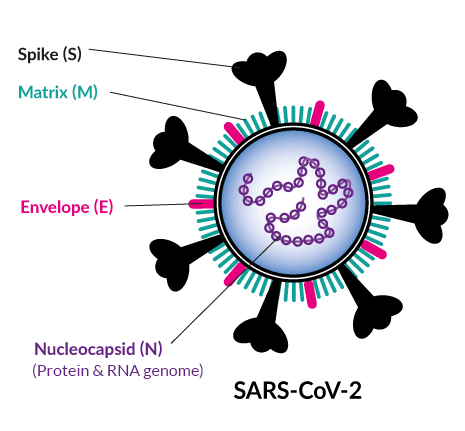
Viruses | Free Full-Text | Potential Molecular Mechanisms of Rare Anti-Tumor Immune Response by SARS-CoV-2 in Isolated Cases of Lymphomas